Click here to sign in with or
Forget Password?
Learn more
share this!
35
Twit
Share
Email
October 10, 2024
This article has been reviewed according to Science X’s editorial process and policies. Editors have highlighted the following attributes while ensuring the content’s credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
by Lucas Johnson, Vanderbilt University
A team of Vanderbilt researchers has released a new benchmarking study that aims to assist scientists in selecting the most effective methods for analyzing spatial transcriptomics (ST) data.
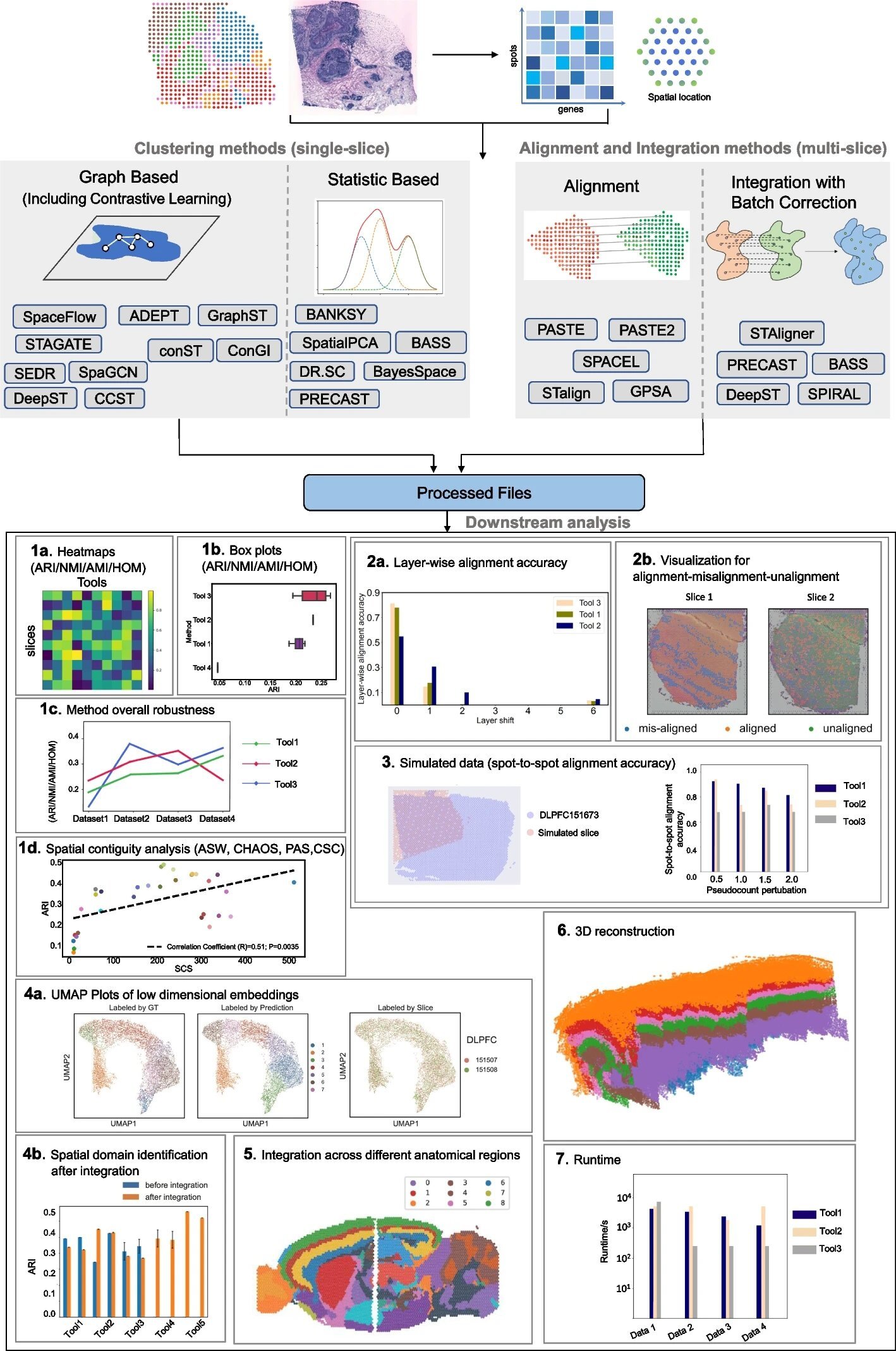
The study led by Xin Maizie Zhou, assistant professor of biomedical engineering and computer science, evaluates computational tools in spatial transcriptomics (ST), a technology used to map gene expression patterns in tissues while preserving spatial context. It was recently published in Genome Biology.
ST involves slicing a tissue sample and placing it on a specially designed slide with spatially indexed barcodes. When the tissue is processed, the ribonucleic acid (RNA) in each specific location of the tissue is captured by these barcodes. After sequencing the RNA, the data can be mapped back to the original tissue locations, allowing researchers to visualize where certain genes are being expressed within the tissue architecture.
Since its widespread use began in 2020, this groundbreaking sequencing technology has revolutionized the understanding of complex tissues. Applications of ST include cancer research and neuroscience, such as mapping gene expression in parts of the brain to understand regional functions or disease mechanisms.
However, the variety of available tools for analyzing data from ST can be overwhelming, making it difficult to choose the right approach for specific research needs.
To address the issue, the Vanderbilt team systematically compared 16 clustering methods, five alignment methods, and five integration methods across a variety of datasets. Their findings offer practical recommendations for researchers working with spatial transcriptomics, helping them to identify tools that best match their research requirements.
“Our goal was to provide a clear and accessible guide for researchers navigating the options available in spatial transcriptomics analysis,” said Zhou, who is among the teaching faculty at Vanderbilt’s trans-institutional Data Science Institute. “We hope this study will be a useful resource for anyone working in this rapidly growing field.”
More information: Yunfei Hu et al, Benchmarking clustering, alignment, and integration methods for spatial transcriptomics, Genome Biology (2024). DOI: 10.1186/s13059-024-03361-0
Journal information: Genome Biology
Journal information: Genome Biology
Provided by Vanderbilt University
Explore further
Facebook
Twitter
Email
Feedback to editors
1 hour ago
0
21 hours ago
0
Oct 10, 2024
0
Oct 9, 2024
0
Oct 9, 2024
0
1 hour ago
3 hours ago
3 hours ago
3 hours ago
15 hours ago
15 hours ago
15 hours ago
15 hours ago
16 hours ago
16 hours ago
6 hours ago
15 hours ago
Oct 7, 2024
Oct 1, 2024
Oct 1, 2024
Sep 25, 2024
More from Biology and Medical
Jan 13, 2023
Jun 6, 2024
Aug 6, 2024
Jun 24, 2024
Mar 15, 2023
Sep 13, 2024
15 hours ago
15 hours ago
16 hours ago
16 hours ago
18 hours ago
18 hours ago
Use this form if you have come across a typo, inaccuracy or would like to send an edit request for the content on this page. For general inquiries, please use our contact form. For general feedback, use the public comments section below (please adhere to guidelines).
Please select the most appropriate category to facilitate processing of your request
Thank you for taking time to provide your feedback to the editors.
Your feedback is important to us. However, we do not guarantee individual replies due to the high volume of messages.
Your email address is used only to let the recipient know who sent the email. Neither your address nor the recipient’s address will be used for any other purpose. The information you enter will appear in your e-mail message and is not retained by Phys.org in any form.
Get weekly and/or daily updates delivered to your inbox. You can unsubscribe at any time and we’ll never share your details to third parties.
More information Privacy policy
We keep our content available to everyone. Consider supporting Science X’s mission by getting a premium account.
Medical research advances and health news
The latest engineering, electronics and technology advances
The most comprehensive sci-tech news coverage on the web